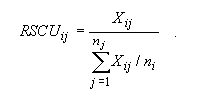 | (3.1) |
In the study of molecular evolution it is often necessary to know some basic statistical quantities such as nucleotide frequencies, codon frequencies, and transition/transversion ratios. The statistical quantities that can be computed by MEGA are discussed in this chapter.
The relative frequencies of the four nucleotides (nucleotide composition) or of the twenty amino acid residues (amino acid composition) can be computed for a specific sequence or for all the sequences used.
Example 3.1 Nucleotide composition of HLA sequences.
---------- Nucleotide composition ----------
All values in per cent (%) except Totals
A T C G Total
HLA-A2 20.8 15.2 29.8 34.2 822
HLA-A3 20.4 14.7 30.2 34.7 822
HLA-All 20.6 14.1 30.5 34.8 822
HLA-AW24 20.9 14.6 30.2 34.3 822
HLA-AW68 20.7 14.8 30.2 34.3 822
All 20.7 14.7 30.2 34.5 4110
For coding regions of DNA, three additional tables are presented for the nucleotide compositions at first, second, and third codon positions. From these tables the G + C content can easily be computed. The amino acid composition can also be presented in a similar tabular form.
There are 64 (43) possible codons that code for 20 amino acids (and stop signals), so an amino acid may be encoded by several codons (e.g., serine is encoded by six codons in nuclear genes). It is therefore interesting to know the codon usage for each amino acid. In MEGA the numbers of the 64 codons used in a gene can be computed either for a specific sequence or for all sequences examined. Four different genetic codes are included; the "universal" code and the mammalian, Drosophila, and yeast mitochondrial genetic codes.
MEGA is also capable of computing Sharp et al.'s (1986) relative synonymous codon usage (RSCU). RSCU is the observed frequency of a codon divided by its expected frequency under the assumption of equal codon usage. That is,
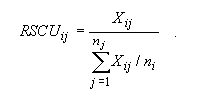 | (3.1) |
Here, Xij. is the number of occurrences of the j-th codon for the i-th amino acid, and ni is the number (from one to six) of alternative codons for the i-th amino acid. This index is useful for knowing the codons that are used more often or less often than expected under the assumption of equal usage.
Example 3.2 Codon frequencies and RSCU values for HLA-A2.
-------- codon Usage --------
Codon Usage Table for HLA-A2
Frequency of codons and relative synonymous codon usage (RSCU)
TTT (F) 0 (0.00) ... TGT (C) 0 (0.00)
TTC (F) 8 (2.00) ... TGC (C) 4 (2.00)
TTA (L) 0 (0.00) ... TGA (*) 0 (0.00)
TTG (L) 2 (0.71) ... TGG (W) 10 (1.00)
.
.
.
GTT (V) 0 (0.00) ... GGT (G) 3 (0.60)
GTC (V) 2 (0.50) ... GGC (G) 7 (1.40)
GTA (V) 0 (o.OO) ... GGA (G) 2 (0.40)
GTG (V) 14 (3.50) ... GGG (G) 8 (1.60)
Total codons scored: 274
'*' indicates a stop codon.
RSCU is given in parentheses.
When two nucleotide sequences are compared, the frequencies of 10 different types of nucleotide pairs can be computed. In MEGA these frequencies are tabulated in the following form.
Example 3.3 Nucleotide pair frequencies for alleles of the HLA-A locus.
------- Observed nucleotide pair frequencies -------
n: total number of nucleotides compared
ns: number of transitional differences
nv: number of transversional differences
nd : ns+nv (total number of nucleotide differences)
Tran- Trans- Identical
sition version pair
AG TC AT AC TG CG AA TT CC GG ns/nv nd n
HLA-A2 vs. HLA-A3 11 5 2 2 5 8 162 117 239 271 0.94 33 822
HLA-A2 vs. HLA-All 11 8 3 4 4 10 161 113 237 271 0.90 40 822
HLA-A2 vs. HLA-AW24 13 11 3 1 5 15 163 113 233 265 1.00 48 822
HLA-A2 vs. HLA-AW68 3 2 2 2 5 Il 167 119 239 272 0.25 25 822
The observed numbers of alignment gaps of different lengths (sites) are useful for studying the distribution of insertions/deletions and for deciding whether all sites containing gaps should be deleted (see section 4.5). In MEGA, the numbers of gaps of length I to 10 can be computed either for each sequence or for all sequences. The numbers of gaps longer than 10 sites are pooled together with the number of gaps of length 10.
Example 3.4 Alignment gap frequencies for HLA sequences.
-------- Alignment Gap Frequencies --------
All entries in the table are the observed number of occurrences
l 2 3 ... >10 Total
HLA-A2 0 0 0 ... 1 1
HLA-A3 0 0 0 ... 1 1
HLA-All 0 0 0 ... 1 1
HLA-AW24 0 0 0 ... 1 1
HLA-AW68 0 0 0 ... 1 1
All 0 0 0 ... 5 5
It is well known that some regions of DNA or amino acid sequences are more variable than others. For example, the control region of mammalian mitochondrial DNA has two hypervariable segments (Kocher and Wilson 1991). One way of detecting such variable regions is to examine the number of variable sites in different segments of the DNA. In MEGA, the numbers of variable sites in overlapping and nonoverlapping segments of equal size can be computed for any segment size (window size). In the output, the numbers of variable sites in overlapping (sliding window) or nonoverlapping segments of a specified size are given along with a histogram.
Example 3.5 Nonoverlapping windows for HLA-A sequence data.
-------- Variability --------
Total number of variable sites: 71
Numbers of variable sites in nonoverlapping segments of size 100
Location
1-1OO | 6 | ******
101-200 | 5 | *****
201-300 | 19 | *******************
301-400 | 10 | **********
401-500 | 7 | *******
501-600 | 13 | *************
601-700 | 5 | *****
701-800 | 5 | *****
801- | 1 | *