
Example Data Files
For this tutorial, we will use example data files that are included
with MEGA. When MEGA-GUI is launched for the first time, it will copy
these example files to your computer in your Documents folder
On Windows: %HOMEPATH%\Documents\MEGA X\Examples
On *nix: ~/Documents/MEGA X/Examples
Demo 1 - Computing a Timetree Using the Reltime Method
Step 1 - Launch MEGA X and set to Prototype mode
Open MEGA X the same way other applications are opened using your operating system
In the main MEGA X window, click the Prototype button to enable the generation of .mao files

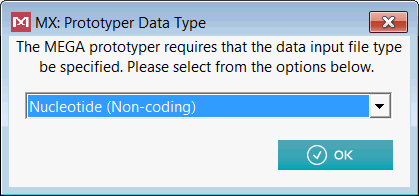
When prompted for the input data type to be used, select Nucleotide (Non-coding) from the drop down list
From the Clocks menu on the main form, select Compute Timetree | Reltime-ML

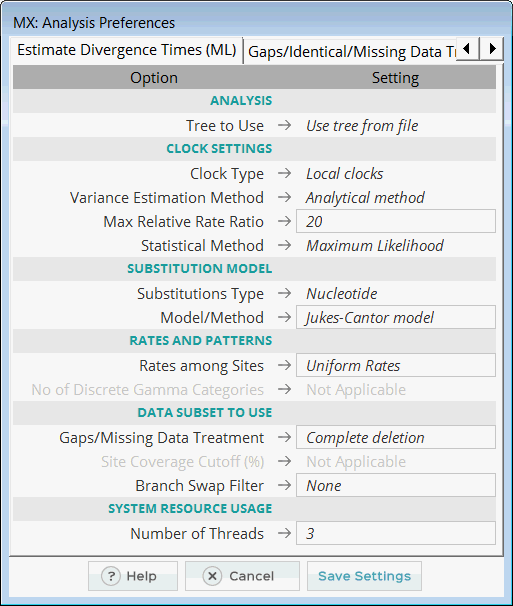
In the Analysis Preferences Dialog, accept the default settings and click the Save Settings... button. When prompted for a location to save
the .mao file, save it in the MEGA X\Examples folder as reltime_ml_nucleotide.mao
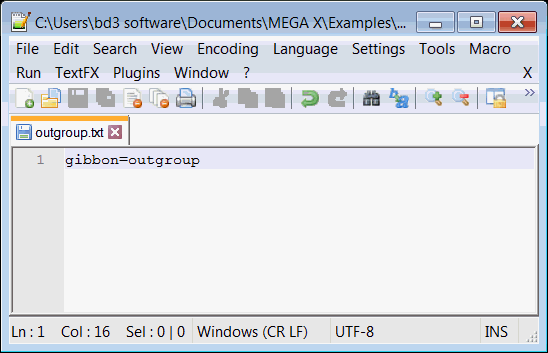
The Reltime analysis requires that we specify an outgroup. Using a text editor, create a text file named outgroup.txt and save it in the
MEGA X\Examples directory. In the text file, add a single line (gibbon=outgroup) which specifies that in our input phylogeny, the outgroup
is comprised of a single taxon named gibbon
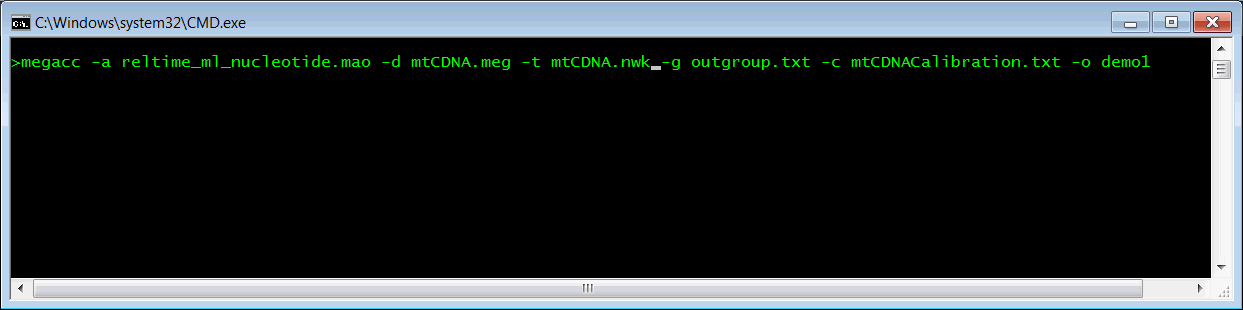
Open a command prompt and navigate to the MEGA X\Examples folder. Launch MEGA-CC by executing the following command:
megacc -a reltime_ml_nucleotide.mao -d mtCDNA.meg -t mtCDNA.nwk -g outgroup.txt -c mtCDNACalibration.txt -o demo1
The Reltime analysis will run and progress updates will be displayed in the command prompt window
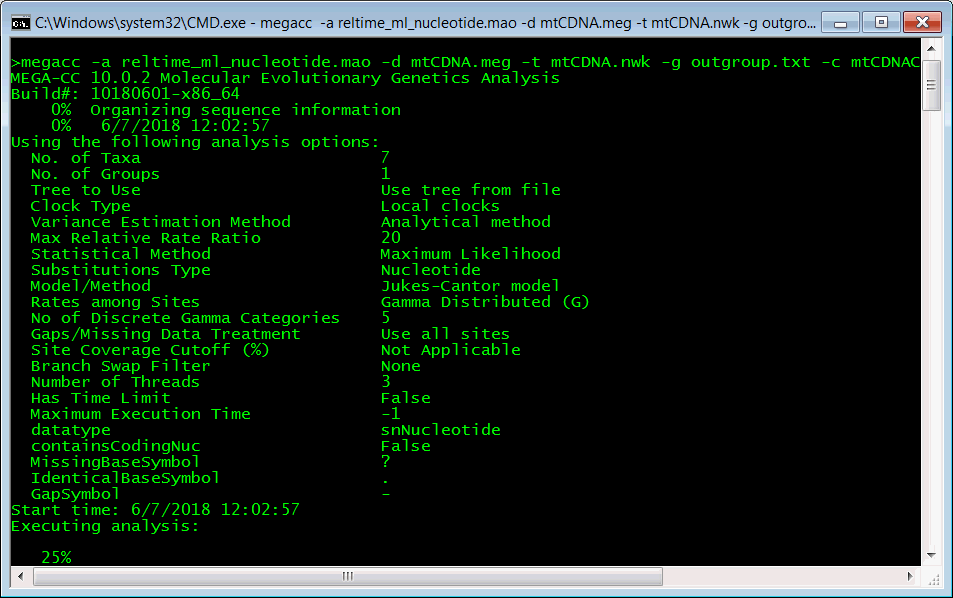
The analysis will produce several output files in the MEGA X\Examples folder:
demo1_exactTimes.nwk
This Newick file gives the timetree scaled according to the estimated divergence times.
demo1_relTimes.nwk
This Newick file gives the timetree scaled according to the estimated relative divergence times.
demo1_nexus.tre
This NEXUS file outputs the timetree in NEXUS format and includes additional information such as divergence time confidence intervals (tip: open this file in the FigTree application for advanced visualization capabilities).
demo1.txt
This text file gives a more detailed representation of the timetree, including relative times, exact times, evolutionary rates, and divergence time std errors.
demo1_summary.txt
This file gives analysis information such as the log likelihood value of the Maximum Likelihood tree, ts/tv ratio, etc...