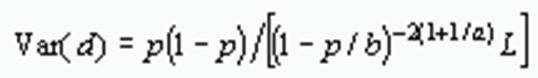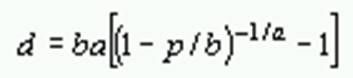
In real data, amino acid frequencies usually vary among the different kinds of amino acids and substitution rates are not uniform among sites. In this case, the correction based on the equal input model gives a better estimate of the number of amino acidsubstitutions than the Poisson correction distance. The rate variation among sites is modeled using the Gamma distribution; for computing this distance you will need to provide a gamma parameter (a).
MEGA provides facilities for computing the following quantities:
Quantity |
Description |
d: distance |
Number of amino acid substitutions per site. |
L: No of valid common sites |
Number of sites compared. |
Formulas used are:
Distance
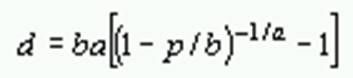
where p is the proportion of different amino acid sites, a is the gamma parameter, gi is the frequency of amino acid i,and
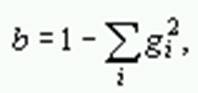
Variance