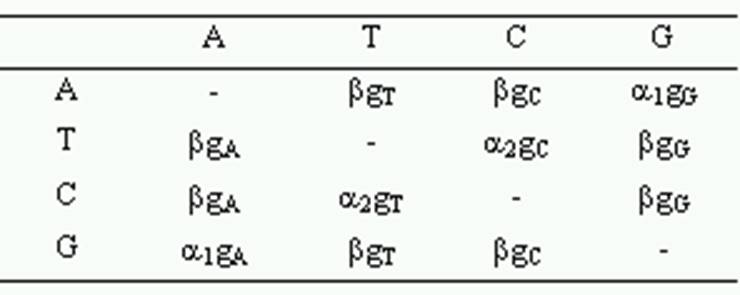
The Tamura-Nei (1993) distance with the gamma model corrects for multiple hits, taking into account the rate substitution differences between nucleotides and the inequality of nucleotide frequencies. In this distance, evolutionary rates among sites are modeled using the gamma distribution. You will need to provide a gamma parameter for computing this distance. When the nucleotide frequencies between the sequences are different, the modified formula (Tamura and Kumar 2002) relaxes the assumption of the substitution pattern homogeneity.
The Tamura-Nei model

MEGA provides facilities for computing the following quantities for this method:
Quantity |
Description |
d: Transitions & Transversions |
Number of nucleotide substitutions per site. |
s: Transitions only |
Number of transitional substitutions per site. |
v: Transversions only |
Number of transversional substitutions per site. |
R = s/v |
Transition/transversions ratio. |
L: No of valid common sites |
Number of sites compared. |
The formulas for computing these quantities are as follows:
Distances
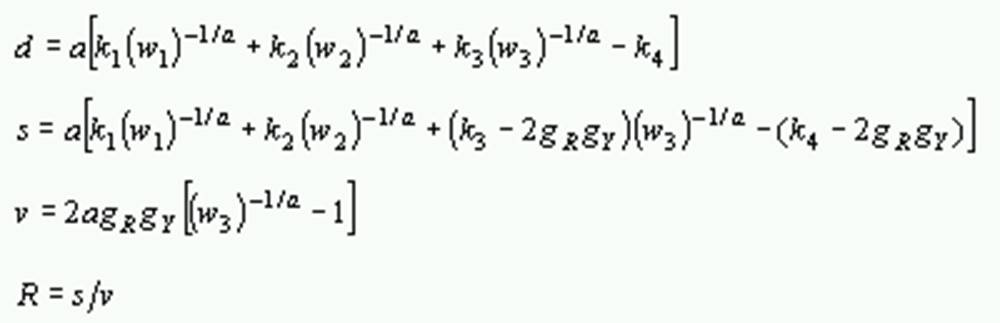
where P1 and P2 are the proportions of transitional differences between nucleotides A and G, and between T and C, respectively, Q is the proportion of transversional differences, gXA, gXC, gXG, gXT, are the respective frequencies of A, C, G and T of sequence X, gXR = gXA + gXG and gXY = gXT + gXC, gA, gC, gG, gT, gR, and gY are the average frequencies of the pair of sequences, a is the gamma parameter and
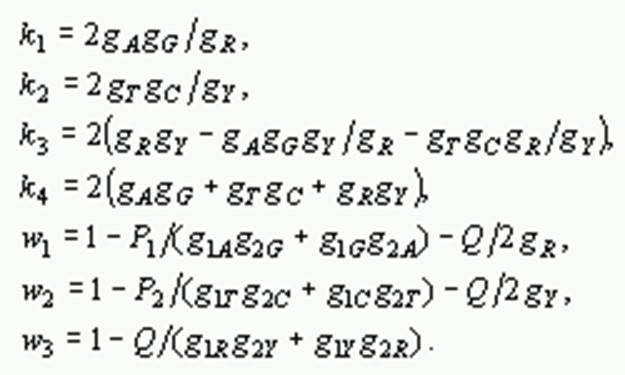
The variances can be estimated by the bootstrap method.