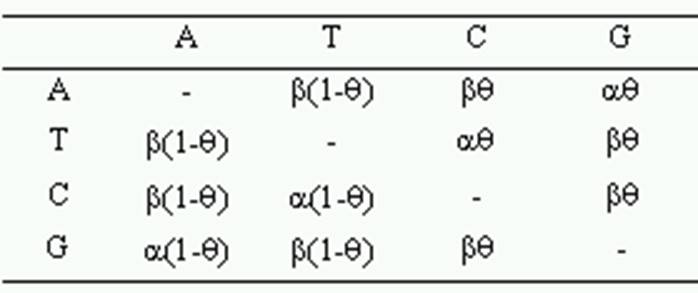
Tamura’s 3-parameter model corrects for multiple hits, taking into account the differences in transitional and transversional rates and the G+C-content bias (1992). Evolutionary rates among sites are modeled using the gamma distribution, and you will need to provide a gamma parameter for computing this distance. When the G+C-contents between the sequences are different, the modified formula (Tamura and Kumar 2002) relaxes the assumption of substitution pattern homogeneity.
The Tamura 3-parameter model

MEGA provides facilities for computing the following quantities:
Quantity |
Description |
d: Transitions & Transversions |
Number of nucleotide substitutions per site. |
s: Transitions only |
Number of transitional substitutions per site. |
v: Transversions only |
Number of transversional substitutions per site. |
R = s/v |
Transition/transversion ratio. |
L: No of valid common sites |
Number of sites compared. |
Formulas for computing these quantities are as follows:
Distances
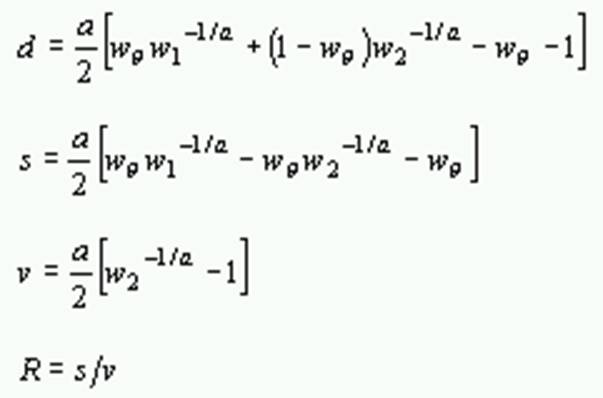
where P and Q are the proportion of sites with transitional and transversional differences, respectively, a is the gamma parameter, and

The variances can be estimated by the bootstrap method.