Thank you for choosing to use MEGA CC (Computational Core) in your research.
The application you are currently used to is MEGA-Proto (MEGA Prototyper). Proto (the application you are running right now) is used to create an MEGA analysis options file, which is required to run an analysis in CC. This allows you to use MEGA visual to specify all of your analysis options, then save a single file to describe your analysis to MEGA CC.
Using MEGA-Proto
MEGA-Proto has the same look and feel as the normal graphical version of MEGA. It produces a MEGA analysis options file (.mao). MEGA-Proto has no computational abilities, it is only made for generating .mao files.
Select input data type.
Nucleotide (non-coding)
Nucleotide (coding)
Protein (amino-acid)
Distance matrix (MEGA format)
Select analysis from menu.
Adjust analysis settings.
Save the MEGA Analysis Options file.
Demo 1
The following example demonstrates how to perform Maximum Likelihood phylogeny reconstruction using MEGA-Proto and MEGA-CC.
For this demo, make sure that you have the M51CC.exe, M51Proto.exe, and example data files located in your working directory as shown. The example data files folder is included in the MEGA-CC.zip download.

Open MEGA-Proto by right-clicking the M51Proto.exe file and selecting Open.
Select the data type of the input data file to be analyzed. For this demo, we will accept the default setting - Nucleotide (non-coding).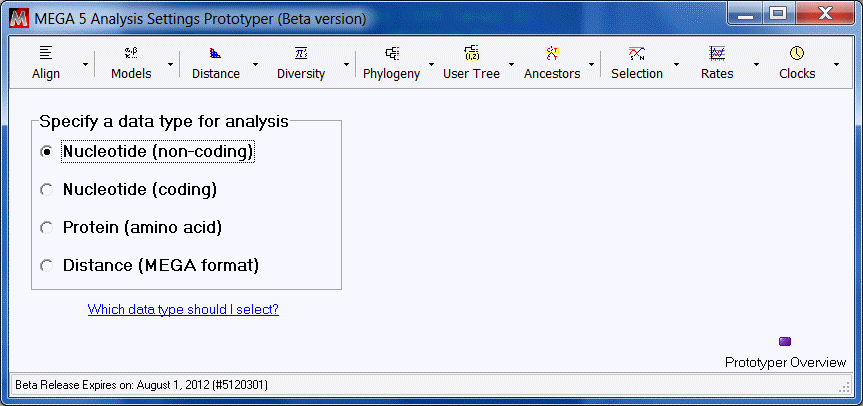
Select Construct/Test Maximum Likelihood Tree
from the Phylogeny menu.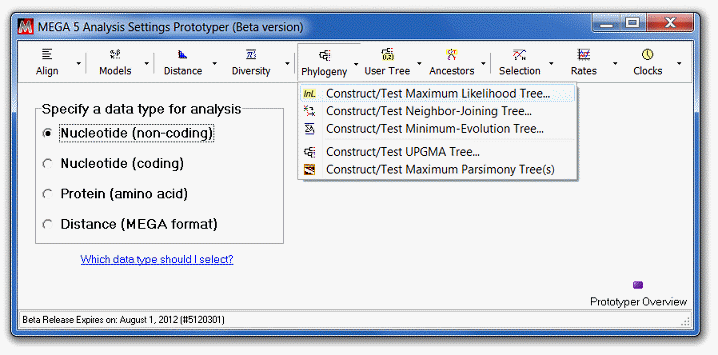
Adjust the analysis preferences to match those shown. 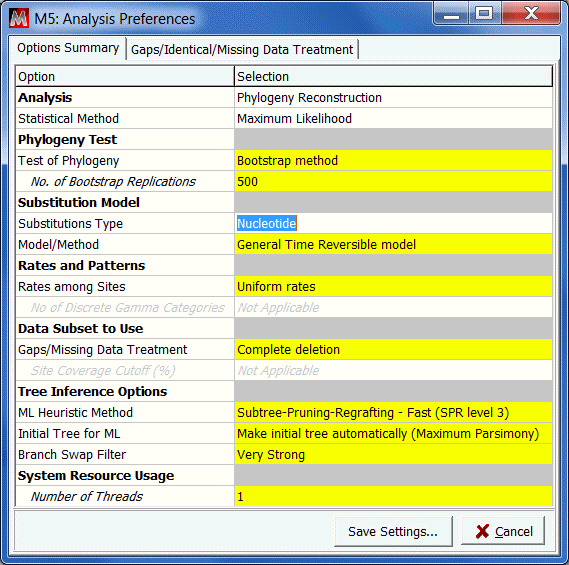
Click the Save Settings
button and save the analysis options file as mlDemo.mao in the current working directory.
Open a command prompt. 
Navigate to the same directory as the M51CC.exe file using the cd command
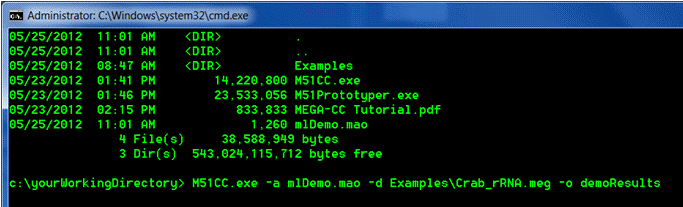
Execute the analysis by calling the M51CC.exe file
M51CC.exe a mlDemo.mao d Examples\Crab_rRNA.meg o demoResults
The analysis will be launched and progress updates will be displayed in the command prompt window.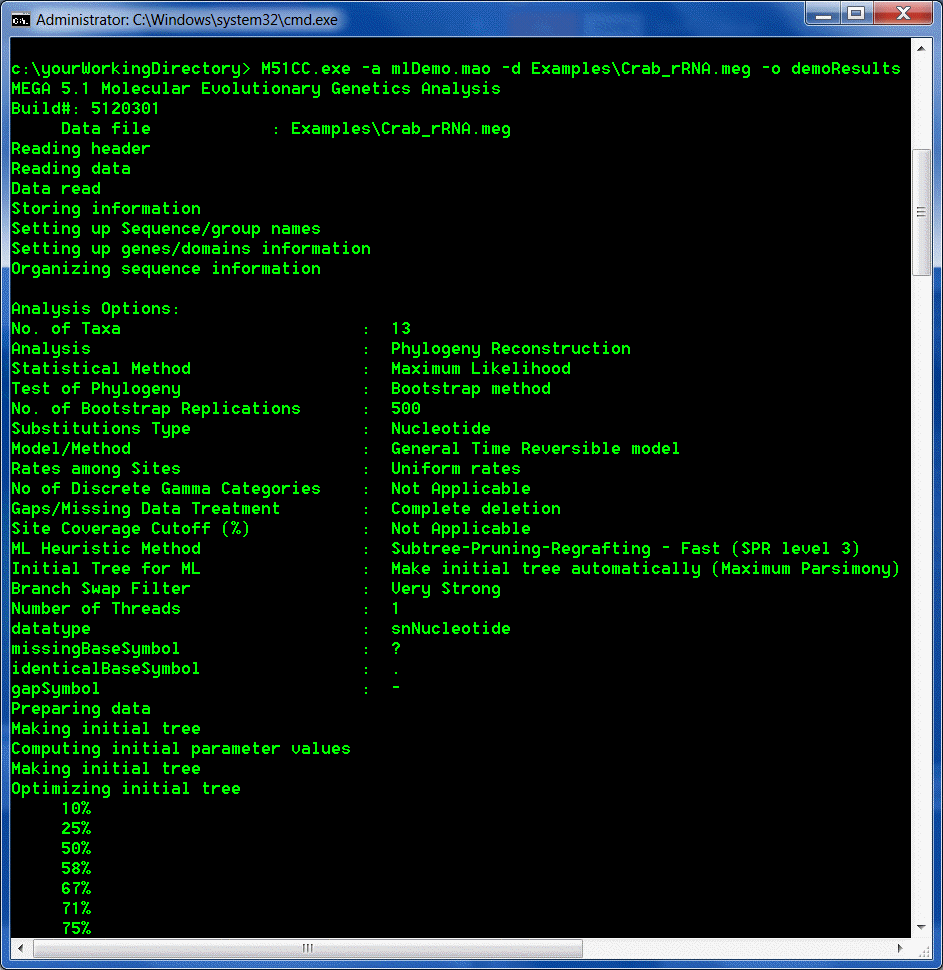
The analysis will produce 3 output files
demoResults.nwk
This Newick file gives the Maximum Likelihood tree found using the
given settings.
demoResults_consensus.nwk
This Newick file gives the bootstrap consensus tree that MEGA
produces from all bootstrap sample trees.
demoResults_summary.txt
This file gives analysis information such as the log likelihood value of
the Maximum Likelihood tree, ts/tv ratio, etc
Demo 2
The following example demonstrates how to use the File Iterator system in MEGA-CC to process multiple input data files using a single analysis options file.
For this demo, make sure that you have the M51CC.exe, M51Proto.exe, and example data files located in your working directory as shown. The example data files folder is included in the MEGA-CC.zip download.

Create a text file named demo2Data.txt which we will use to specify multiple alignment files for ML phylogeny inference.
In this file, add the full paths to the Crab_rRNA.meg and Drosophila_Adh.meg example files. 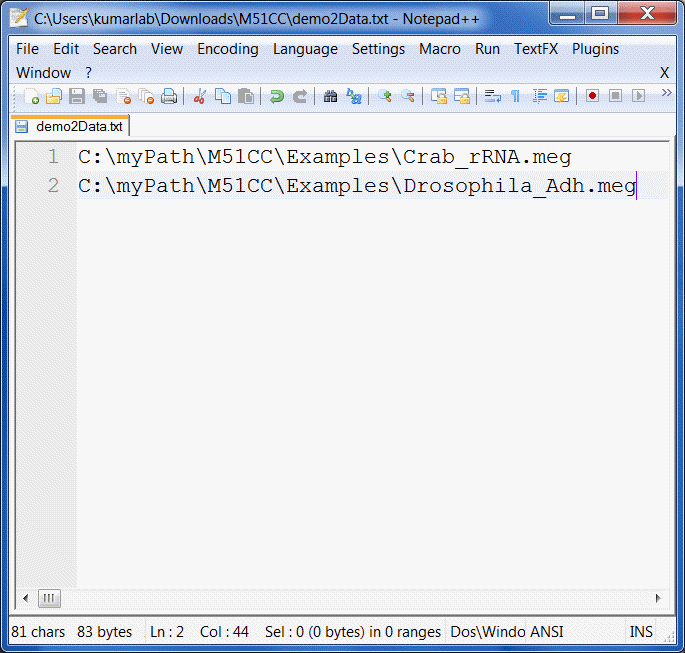
From a command-line prompt, call MEGA-CC as follows:
M51CC.exe a mlDemo.mao d demo2Data.txt
The analyses will be launched sequentially and progress updates will be displayed in the command prompt window.
Wait for all analysis to finish.
The analysis will produce output files for each input data file. In this example, the same analysis options were used for each alignment file
CC
Installation
There is no need to actually install MEGA-CC or MEGA-Proto. MEGA CC is a command line application and must be opened via the command prompt (Start -> All Programs -> Accessories -> Command Prompt). MEGA Proto can be opened like any other application, by double clicking on it.
Initially you should copy all of the files from the zip file to a separate directory, where you will be able to easily access it. (i.e. My Documents\MEGA-CC\).
MEGA-CC Input Files
MEGA Analysis Options file
Specifies the calculation and desired settings.
Created using MEGA-Proto.
Has a .mao file extension.
Data file (one of the following)
Multiple sequence alignment in MEGA or Fasta format.
Distance matrix in MEGA format.
Unaligned sequences in Fasta format (for alignment only).
Tree file (required for some analyses)
Newick file format.
MEGA-CC Output Files
In general, two output files are produced
Calculation-specific results file (Newick file, distance matrix, ).
A summary file with additional info (likelihood, SBL, ).
Some analyses produce additional output (bootstrap consensus tree).
Output directory/filename
Default is the same location as the input data file.
Specify an output directory and/or file name using -o option.
If no output filename is specified, MEGA-CC will assign a unique name.
Errors/warnings
If MEGA-CC produces any errors or warnings, they will be logged in a file named M5ErrorLog.txt in the same location as the results. Always check for the existence of this file.
Running MEGA-CC
Easiest to run using command-line or batch scripts:
M51CC.exe a options.mao d alignment.meg o outFile
Can also be run using custom scripts (Perl, Python, ):
exec(M51CC.exe a options.mao d alignment.meg o outFile);
Integrated File Iterator system can process multiple files without the need for using scripts (see Demo2 below)
In addition, other applications can launch MEGA-CC:
status = CreateProcess(M51CC.exe );
To see a list of available command options, call M51CC.exe from a command-line prompt with the h flag.