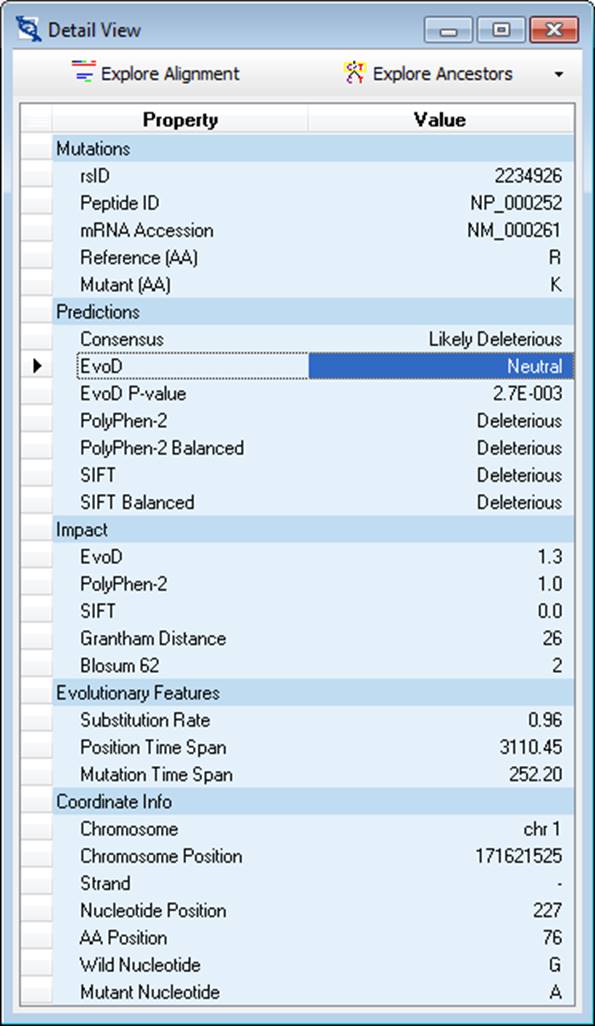
The Mutation Detail View window displays all available information for the currently active record (selected in the Mutation Explorer window). Additionally, this window provides access to the 46-species reference alignment for the given gene as well as the ability to infer ancestral alleles using the Maximum Likelihood (ML) or Maximum Parsimony (MP) methods.
When the Explore Alignment button is clicked, MEGA-MD will retrieve the 46-species reference alignment from the MEGA-MDW server and display it in the Sequence Data Explorer, from where it can be exported or further exploration can be done.
When the Explore Ancestors button is clicked, the choice of ML and MP methods are presented. If the ML approach is selected, the Analysis Preferences Dialog is displayed from which the analysis can be launched with custom settings (e.g. substitution model, distribution of rates, etc…). If the MP approach is selected, the analysis is launched immediately as not custom settings are available for this method. When the analysis is completed, the reference topology will be displayed in the Tree Explorer along with inferred ancestral alleles for the amino acid site designated earlier.
