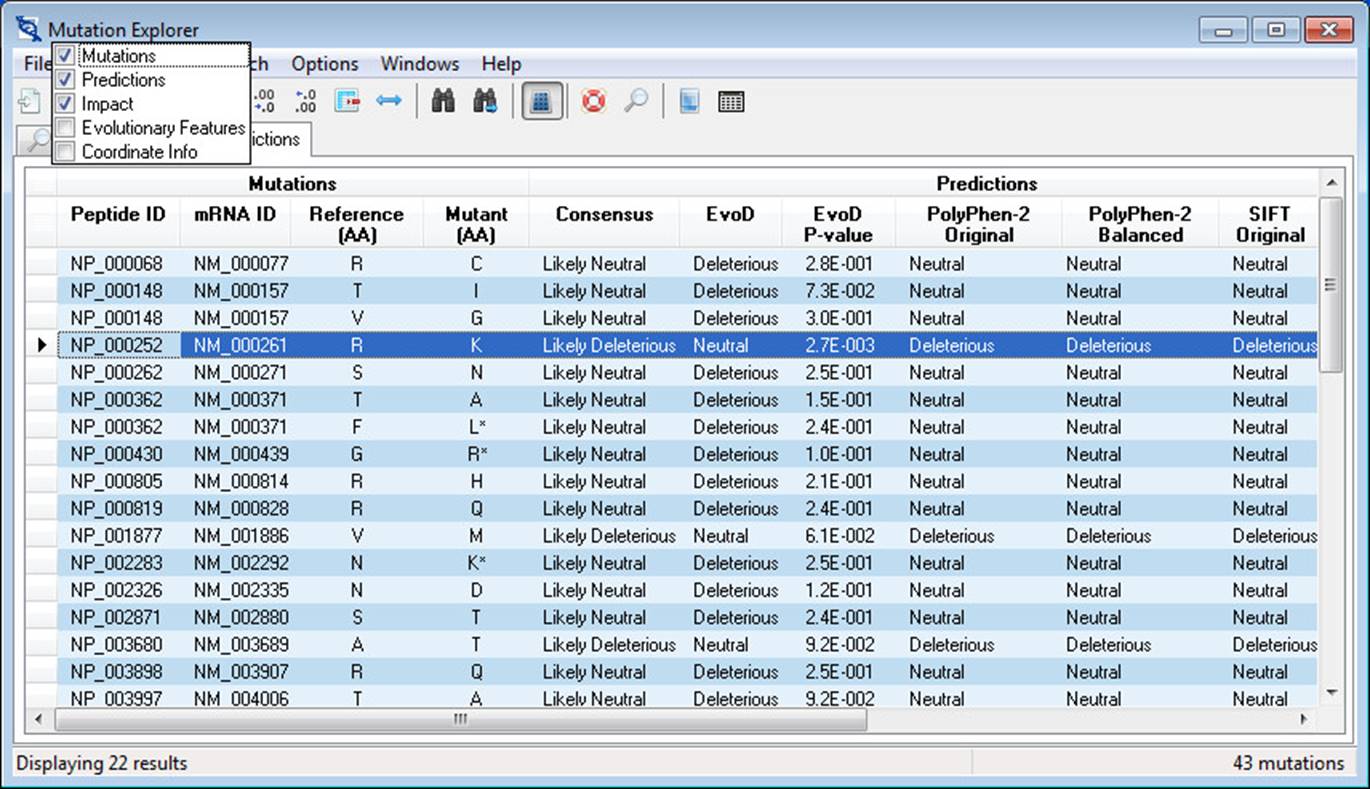
The Predictions tab displays all prediction data retrieved from the MEGA-MDW server in a list view. Complete information for the currently active record is displayed in the Mutation Detail View. Columns of data are banded together into categories:
· Mutations – identifiers as well as mutant and reference alleles are given here. Note – mutant amino acids that are appended with an asterisk (*) have multiple rows returned by the MEGA-MD server, each row indicating a mutation at the nucleotide level (look to the Coordinate Info band to see nucleotide change).
· Predictions – consensus, EvoD, PolyPhen-2, and SIFT predictions are given here. Where both the original and balanced predictions are given for PolyPhen-2 and SIFT (balanced predictions are described in Liu and Kumar 2013).
· Impact – the impact scores for EvoD, PolyPhen-2, and SIFT predictions are provided along with the Grantham distance and Blosum62 value.
· Evolutionary Features (hidden by default) – substitution rate, position time span, and mutation time span are displayed (see below for a description of how to display this band).
· Coordinate Info (hidden by default) – additional coordinate information is shown here, including chromosome, strand, nucleotide position, amino acid position, wild nucleotide, and mutant nucleotide (see below for a description of how to display this band).
To toggle on/off the display of a given band, click on the indicator button which is located to the far left in the band headers row. A popup menu will appear from which bands can be selected/deselected. Often times when changing the display of bands, column widths will change in undesirable ways. To remedy this, you can execute the Best-fit Columns action by clicking Format->Resize columns to best-fit or clicking the toolbar button. Alternatively, columns widths can be adjusted by dragging their header edges.
The toolbar and main menu provide access to several actions for importing/exporting data, formatting the view, sorting, text search, and setting view options.
